Dr. Mochly-Rosen and grad students publish COVID-19 variant work
Posted on June 11th, 2021
Among the scientists turning their attention to investigate COVID-19 this past year, Dr. Daria Mochly-Rosen recruited two first year graduate students and rapidly produced three studies on SARS-CoV-2 variants. After first posting the studies to bioRxiv in December and January, two of the papers have now been published in peer-reviewed journals.
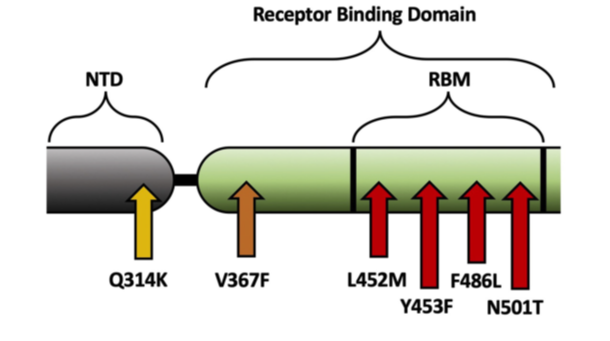
The team’s study investigating why two SARS-CoV-2 variants are more transmissible was recently published in PloS One. The team found that mutations in the SARS-CoV-2 spike protein could increase the sensitivity of the protein to cleavage by proteases, or increase the chances of the spike protein to be in the open, active conformation, both of which expose the receptor-binding domain, better enabling the virus to bind to the ACE2 receptor and enter the body.
The team’s work examining mutations that developed as the SARS-CoV-2 virus moved between mink populations and humans was published in Infection, Genetics and Evolution in early May.
SPARK wrote about the grad students’ perspective working on these projects with Mochly-Rosen in our Bridging the Gap blog. The students, Suman Pokhrel and Ben Kraemer, were doing rotations in Mochly-Rosen’s lab. They were concerned about reports of mutations in SARS-CoV-2, and used computational methods to examine coronavirus genomic sequences deposited as open-source data in GISAID, with the hope that their work could aid development of vaccines and therapeutics to stop COVID-19 transmission.
It was an incredible achievement for the students, said Mochly-Rosen, and a unique, surreal experience for students starting graduate school in the middle of a pandemic.
The team’s third study investigating natural SARS-CoV-2 variants has been accepted for publication in a peer-reviewed journal. That study looked at variants that arose naturally as the virus spread around the world, and identified regions in the spike protein with limited mutations that could be targeted with prophylactics and therapeutics.
Permalink: https://sparkmed.stanford.edu/mochly-rosen-publish-studies-covid-19-variants/
